PhysiCell
I am currently working with Dr. Paul Macklin in Intelligent Systems Engineering, SICE, at IU.
Our primary focus is on developing and supporting PhysiCell, related projects,
and the user communities surrounding them.
Visualization
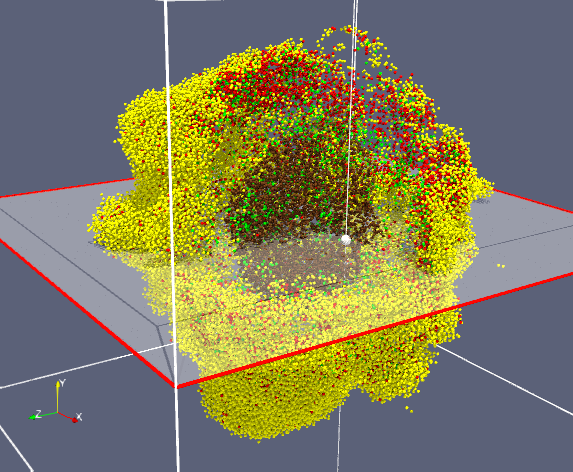
Snapshot of data from a tumor simulation visualized with ParaView. Interactive cutting planes
reveal a necrotic core.
nanoBIO
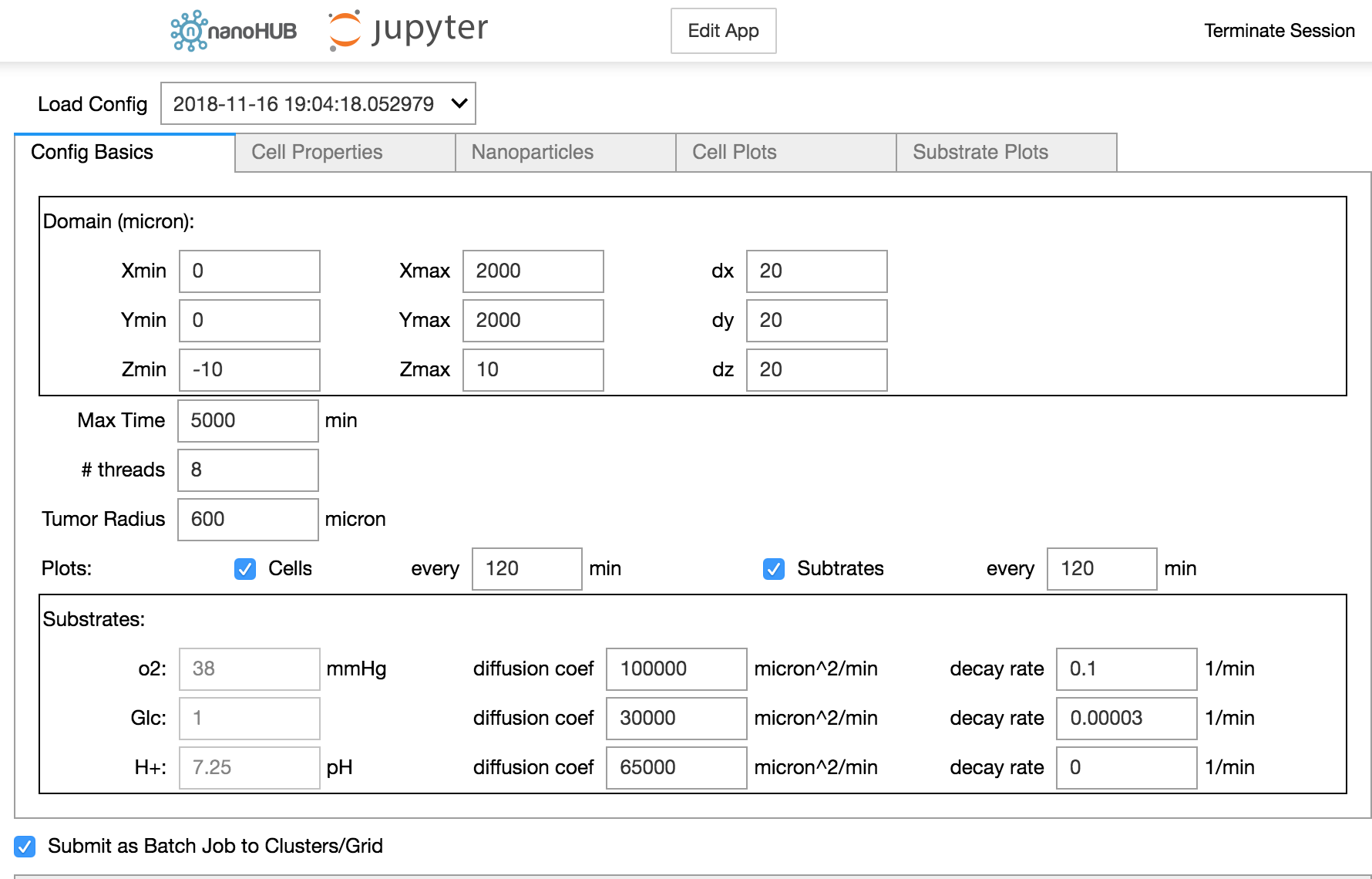
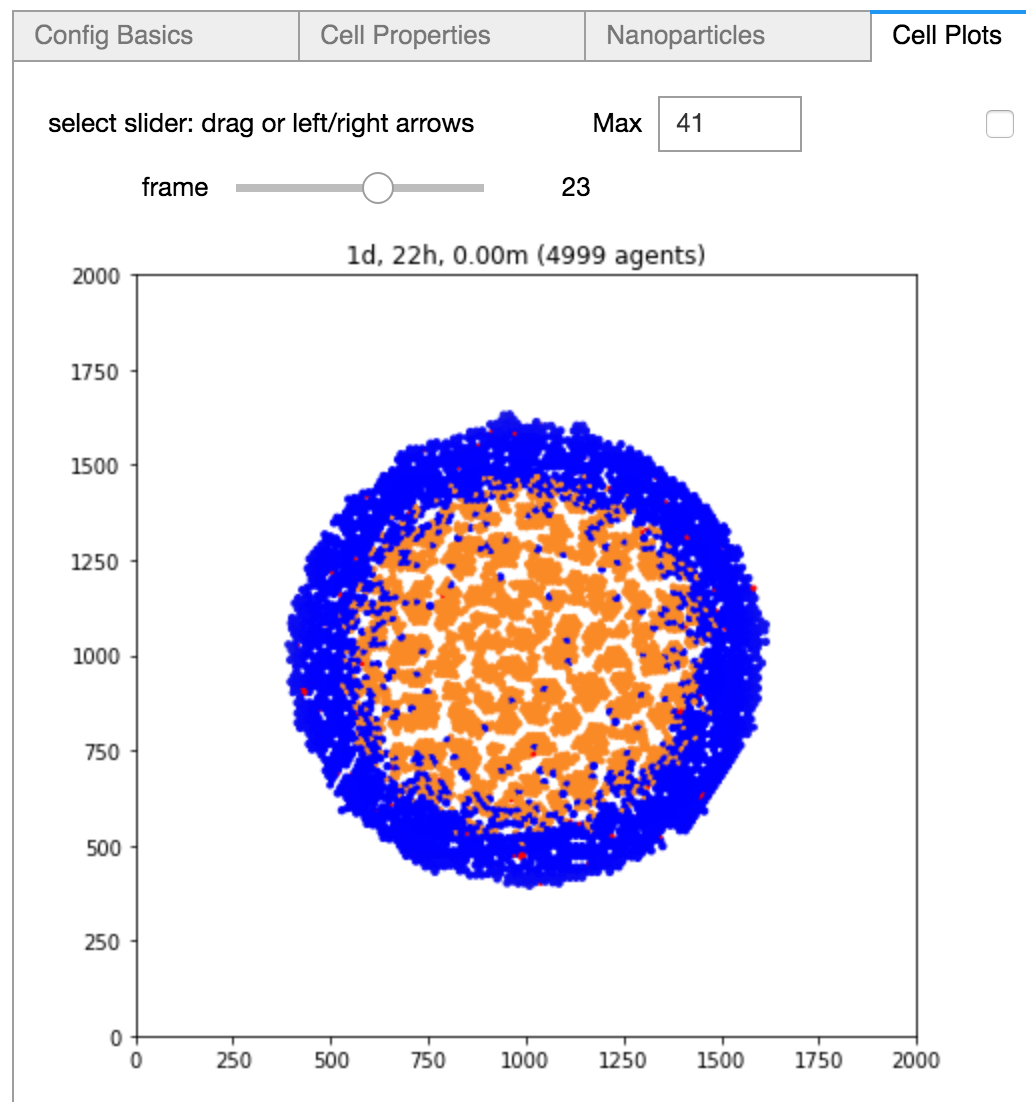
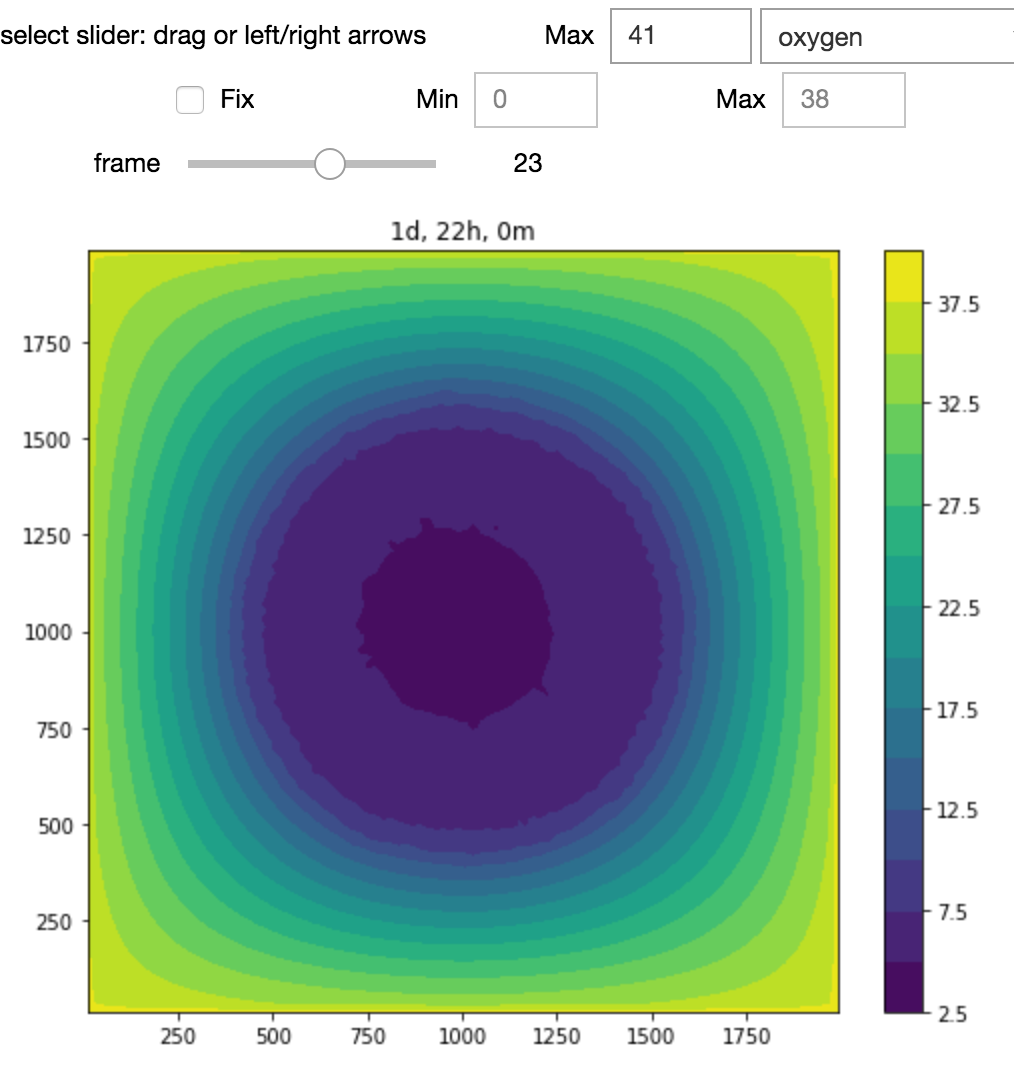
Jupyter notebook GUI for our nanoBIO (PhysiCell-based) nanoparticle application running on nanoHUB.
Low-cost computers
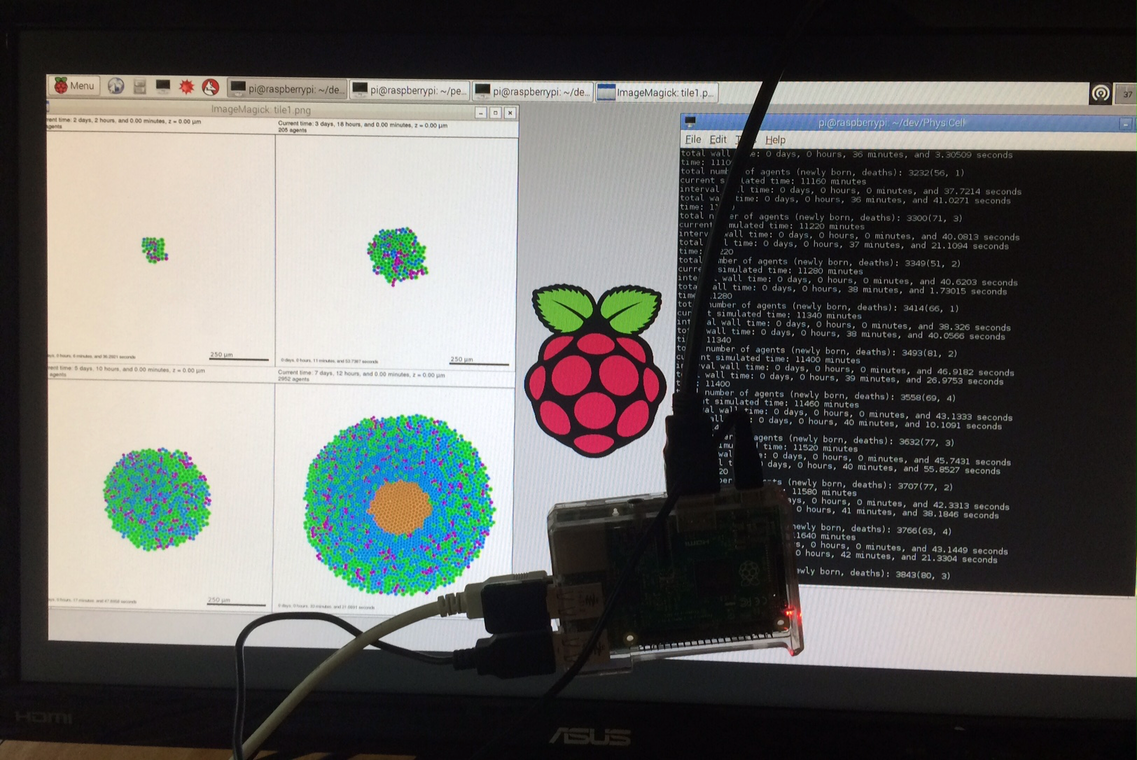
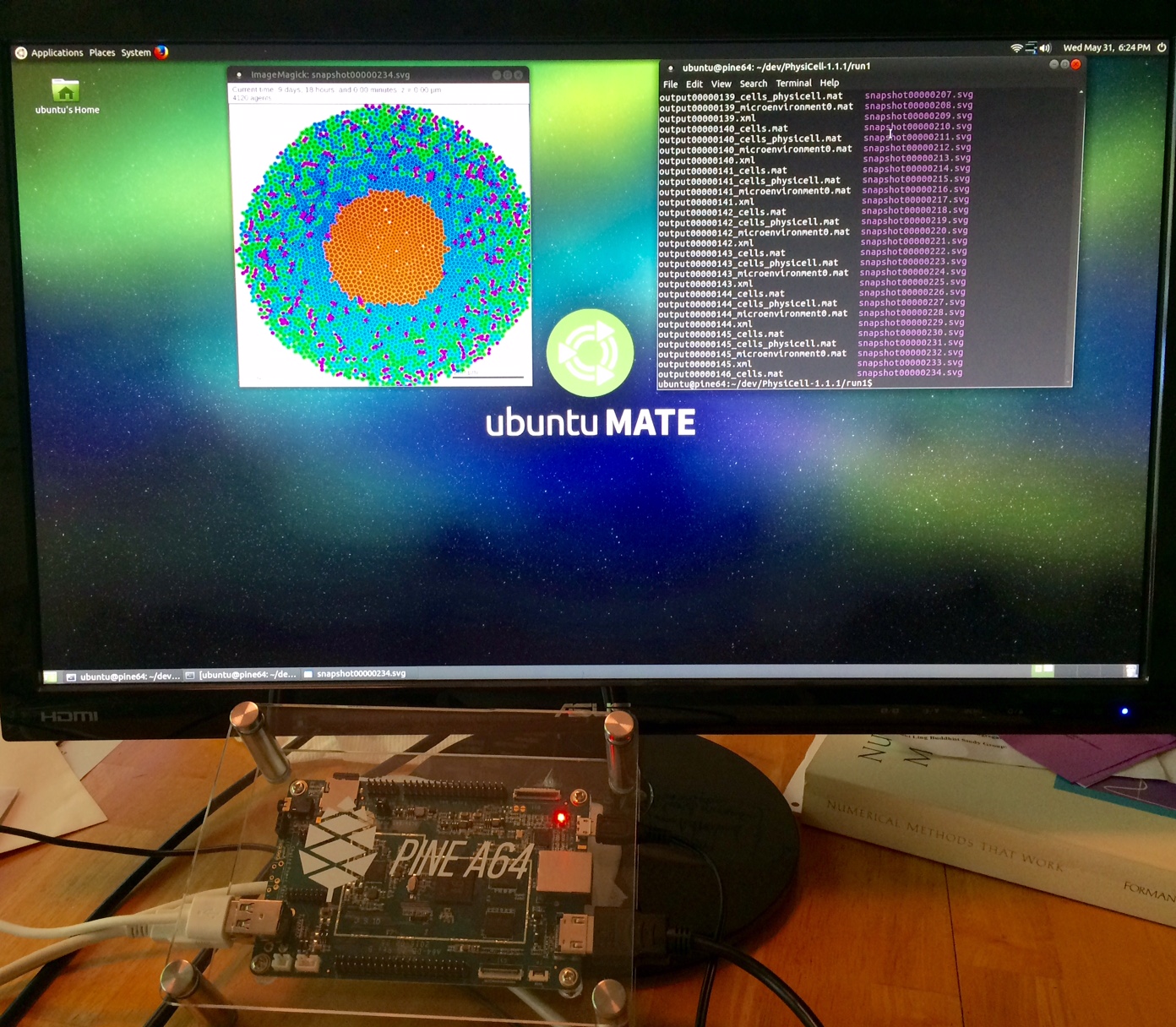
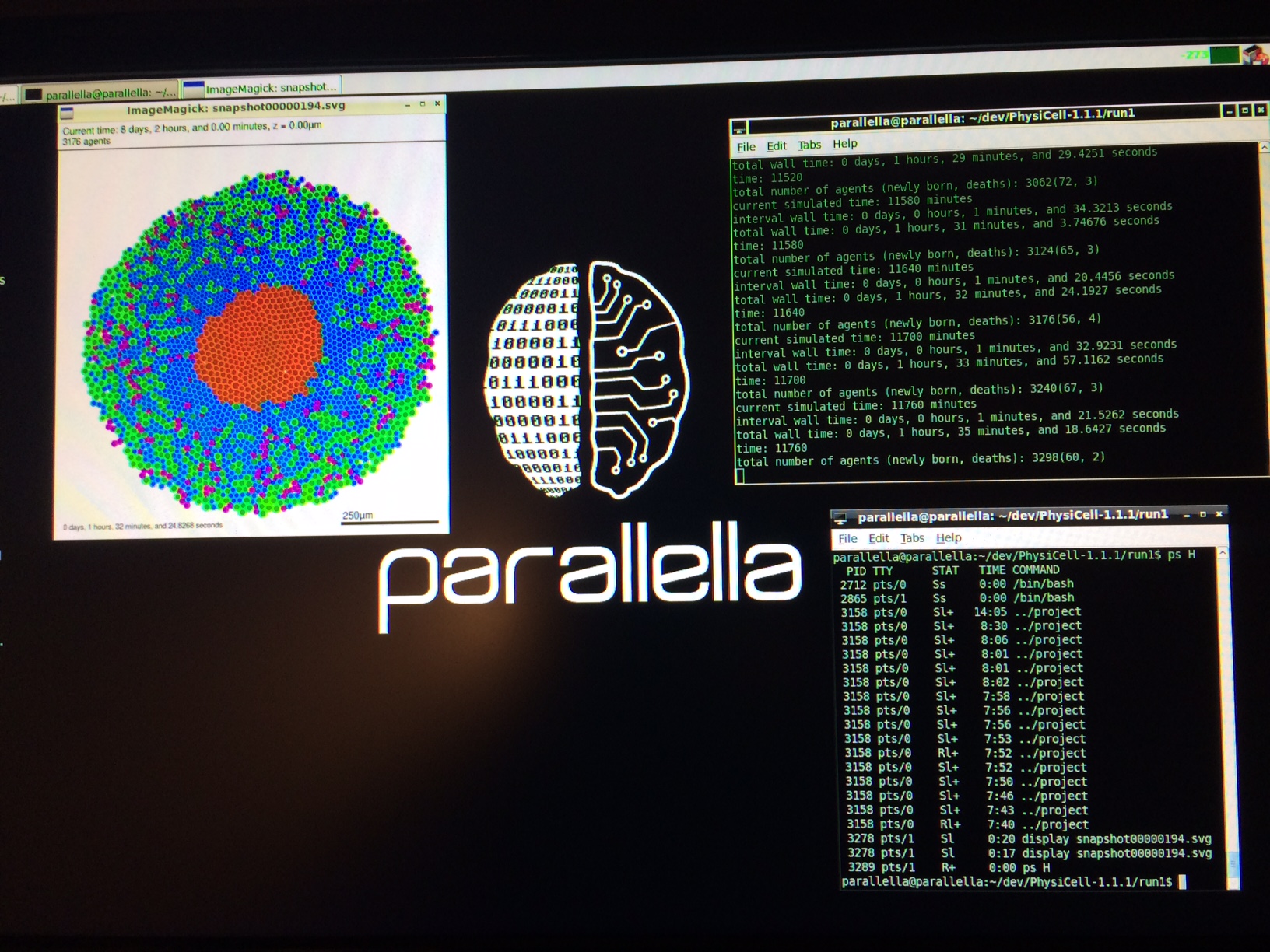
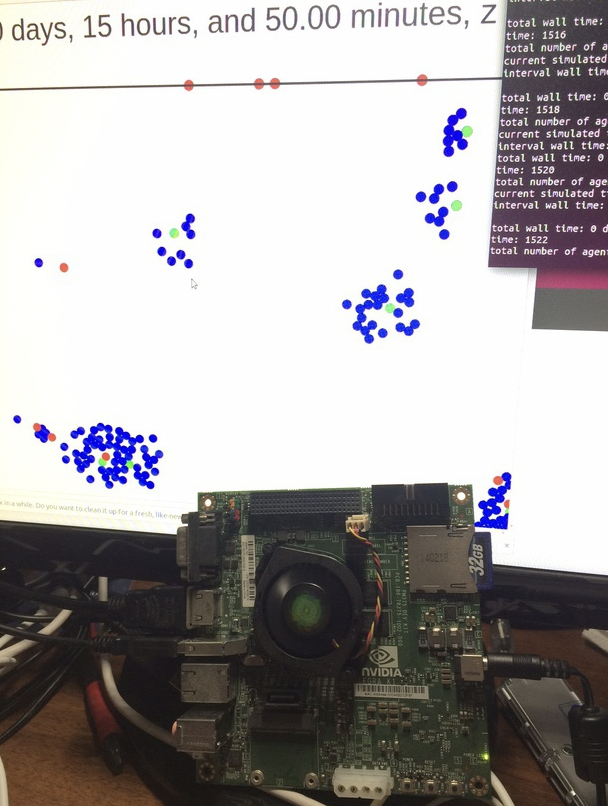
We have demonstrated that PhysiCell also runs on several inexpensive computing platforms.
References
- Ghaffarizadeh, A., Heiland, R., Friedman, S.H., Mumenthaler, S.M., and Macklin, P., PhysiCell: an open source physics-based cell simulator for 3-D multicellular systems. 10.1371/journal.pcbi.1005991
- High-throughput cancer hypothesis testing with an integrated PhysiCell-EMEWS workflow.
Jonathan Ozik, Nicholson Collier, Justin Wozniak, Charles Macal, Chase Cockrell, Samuel Friedman, Ahmadreza Ghaffarizadeh, Randy Heiland, Gary An, Paul Macklin.
https://doi.org/10.1186/s12859-018-2510-x
- PhysiBoSS: a multi-scale agent-based modelling framework integrating physical dimension and cell signalling. Gaelle Letort, Arnau Montagud, Gautier Stoll, Randy Heiland, Emmanuel Barillot, Paul Macklin, Andrei Zinovyev, Laurence Calzone. https://doi.org/10.1093/bioinformatics/bty766
- https://news.iu.edu/stories/2017/09/iub/releases/21-engineered-nanobio-hub.html





