
Snapshot of the CompuCell3D desktop application showing a liver hepatocyte simulation.
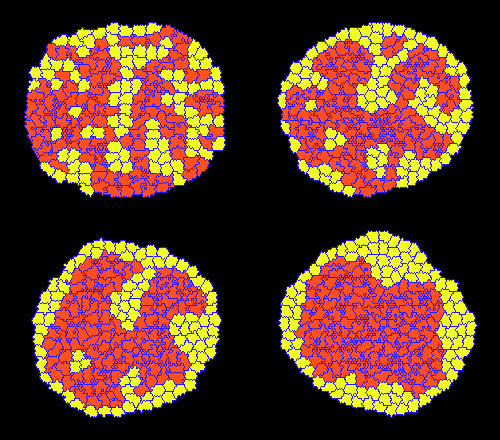
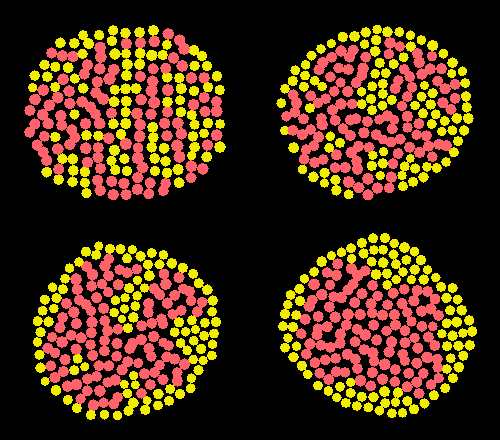
Classic cell sorting simulation: lattice and glyphs visualization.

Snapshots from a wet foam simulation demonstrating coarsening. (movie1, movie2)

3D printed model showing necrotic core (purple) and vasculature (white spheres along vessels are simply artifacts to hold together disjoint pieces and offer support).
Visualizing Cells and their Connectivity Graphs for CompuCell3D
R. Heiland, M. Swat, J. Sluka, B. Zaitlen, A. Shirinifard, G. Thomas, A. Lumsdaine, and J. Glazier
IEEE BioVis Symposium, Oct 2012
Workflows for Parameter Studies of Multi-Cell Modeling
Randy Heiland, Maciek Swat, Benjamin Zaitlen, James Glazier, Andrew Lumsdaine
HPC 2010
Multi-Cell Simulations of Development and Disease Using the CompuCell3D Simulation Environment
Maciej Swat, Susan D. Hester, Ariel I. Balter, Randy W. Heiland, Benjamin L. Zaitlen, James A. Glazier
book chapter in Systems Biology Series: Methods in Molecular Biology Vol. 500 Maly, Ivan V. (Ed.) 2009, ISBN: 978-1-934115-64-0
The cell behavior ontology: describing the intrinsic biological behaviors of real and model cells seen as active agents.
Sluka, J. P., Shirinifard, A., Swat, M., Cosmanescu, A., Heiland, R. W., & Glazier, J. A. (2014).
Bioinformatics 30 (16): 2367–74.